 Home > Achievement > 2022 Research Project List > Molecular Marker-Assisted Tomato Disease Resistance Breeding Selection Model Construction and Technology Development Home > Achievement > 2022 Research Project List > Molecular Marker-Assisted Tomato Disease Resistance Breeding Selection Model Construction and Technology Development |
Molecular Marker-Assisted Tomato Disease Resistance Breeding Selection Model Construction and Technology Development
Although Taiwan's tomato varieties are popular in the international market, with the continuous introduction of new varieties, the market life of tomato varieties is getting shorter and shorter, and the pressure on variety research and development is becoming heavier. The R&D strategy of using molecular markers for specific gene screening to confirm whether the target gene of the contributing parent is introduced into the reincarnating parent is gradually being used; in this study, by cooperating with the industry, applying molecular marker prospect and background genetic analysis, targeting tomato wilt disease resistance gene I -3 Introduction and backcross breeding collaboration, integrate foreground and background selection strategies, and establish a set of molecular collaborative breeding models. By collecting the public tomato SNP database and genome sequence, designing SNP markers, 32 groups of tomato background genetic analysis molecular markers have been developed and completed; the establishment of main prospects and selection of I-3 genotype analysis SCAR markers, in order to improve the screening of a large number of backcross descendants Efficiency, convert SCAR markers into KASP molecular markers, assist seedling screening of BC1F1 single plants, confirm the introduction of disease resistance genes into single plants, enter the BC2F1 selection generation, and improve breeding efficiency.
 ▲Fig.1 |
 ▲Fig.2 ▲Fig.2 |
|
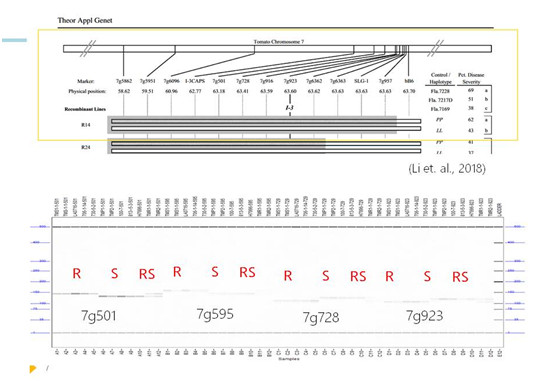 ▲Fig.3 |
