- The application of Single Nucleotide Polymorphism (SNP) markers in identification of resistance in tomato and hybrid seed purity in watermelon.
- Establishment and Research of pathogen detection and protection techniques of propagation Seed and Seedling pathogens
- The promotion of seed nursery joint marketing information platform
- Establishment of Detection Techniques for Seedborne Pathogens of Tomato
- Metabolic Virulence Determinant DNA detection of Bacterial spot of Tomato
- Develpoment of potyvirus resistant germplasm for melon
- Study on in vitro micropropagation of traditional Chinese medicines and edible bamboo crops
- The establishment and application of specific molecular markers for Fusarium crown and root rot resistance gene in tomato.
- Construction of accreditation systems for plant seeds and seedlings
- Study of organic materials for disease control applied to seed coating
- Intensification of Plant New Variety DUS Testing Technology and Research of Genetic Resources Management of the Potential Crop in Taiwan
- Export potential bulb flowers breeding and establishment the propagation system
- Breeding and Critical Techniques Development of Orchids.
- Breeding and evaluation of cucumber’s rootstock on efficient water resources utilization
- Development of cereal residues made plug trays producing techniques and the effects on seedling production.
- Study on organic culture of eggplant
- Breeding for disease resistance in tomato and high quality eggplant.
- Study on improvement of melon vegetable varieties
- Study to make Industrial value chain more competitive for expert in the seedlings industry
- International competitiveness of the breeding of quality varieties in papaya
- Construction of Integrated Plant Seedling Multi-detection Services Platform
- Study of inspection acts for imported genetically modified feed and industrial applications traceability and export agricultural products
- Establishment of transgenic crops of high-performance monitoring system
- Study on the Building of the talent certification Comformed to Occupational Competency in Vegetable Seed Industry
- The training performance evaluation and the analysis of working in agriculture by seedling standardized course in the farmers’ academy
- Enhancing industry connection of seed testing and international cooperation of ISTA
- Interaction of techniques and quality assurance system on seedborne pathogens detection of plant seeds for export
- Cacao breeding and development of vegetative propagation techniques
- Development of breeding and commercialization mode of Vandeae
 Home > Achievement > Research Project Summary Report > 2017 year > The application of Single Nucleotide Polymorphism (SNP) markers in identification of resistance in tomato and hybrid seed purity in watermelon. Home > Achievement > Research Project Summary Report > 2017 year > The application of Single Nucleotide Polymorphism (SNP) markers in identification of resistance in tomato and hybrid seed purity in watermelon. |
The application of Single Nucleotide Polymorphism (SNP) markers in identification of resistance in tomato and hybrid seed purity in watermelon.
In this study, the tomato resistance-related gene sequences of Sw-5 ,Tm-1,Tm-22 ,Ph-2 and Ph-3 genes were used to design SNP primers. The resistance and susceptible tomato materials were screened by using PCR and real-time PCR. Then the screening results showed that these tomato materials could be divided into three resistance genotypes in Sw-5 ,Tm-1 ,Tm-22, Ph-2 and Ph-3 genes. In addition, we had done the molecular detection and field traits with two-hybrid tomato F1 materials from KNOWYOU SEED company. We selected 20 single plants with resistance genes and good characteristics based on the analyzed results. Seven single plants of them were licensed to three companies. Analyzed 6,784,860 SNPs from public genome data of watermelon, 2,156 SNPs whose minor allele frequency (MAF) were more than 0.47 were selected and designed to 857 allele specific markers among which 140 AS markers were synthesized and tested. The results showed that 63 AS markers are polymorphic among the tested hybrid varieties as hybrid seed purity test markers. It was expected that these markers could be widely used in other watermelon varieties.
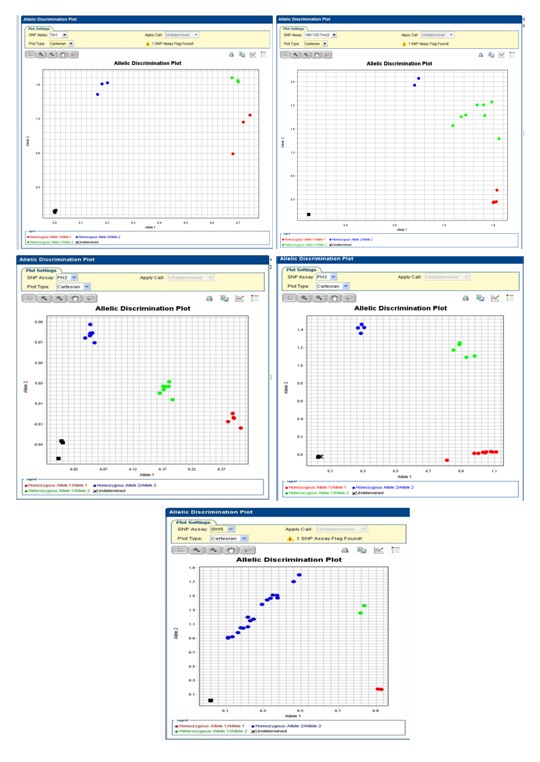
▲Fig. 1 The SNP molecular markers linked to resistant
genes of tomato mosaic virus, Phytophthora infestans
and tomato spotted wilt virus developed by TSIP.
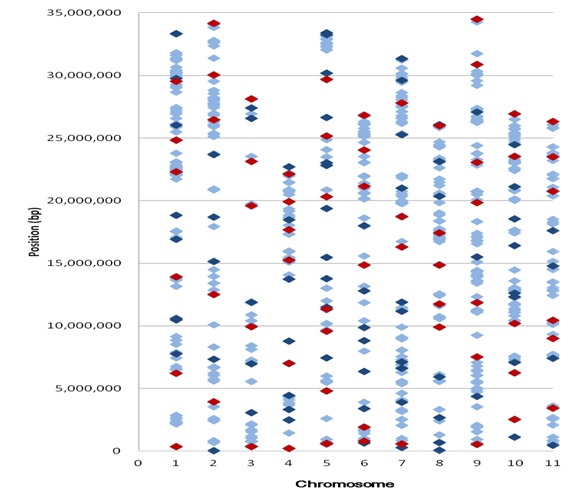
▲Fig. 2 Distribution of SNPs designed as the allele-specific (AS)
markers on the genome of watermelon. The light blue spots were
857 SNPs initially selected; the dark blue and red spots were the
140 SNP (AS) markers tested in the electrophoresis analysis; and
the red spots were the 63 polymorphic candidate SNPs developed
in this study.
