- Verification Of onion Healthy Seedling Cultivation Adaptation Technology in Pingtung and Tomato High Temperature and Drought Technology under Heavy Rainfall Situation in Tainan
- Carbon Footprint Assessment of Local Dried Shiitake Mushrooms
- The Promotion of Conservation systems and the assessment of applications for indigenous heirloom foxtail millet(Setaria italia)
- Establishment and Verification of the Optimal Irrigation Mode for Melon in Protected Facility
- Establishment of Important Virus Detection Method for Grape in Taiwan
- Restablishment of Domestic Seed Corn and Sorghum Production System
- Construction of Environmental Verification System and Standards for Cucurbit Vegetables Nurserie
- The Promotion of Healthy Germplasm Conservation and Propagation Systems for Taro and Ginger
- Establishment of Techniques for Phytoplasma Detection and Classification and Investigation on Phytoplasma Pathogen in Seeds of Important Crops
- Study on Tomato Rootstock Resistance to Bacterial Wilt and Implementation of Field Management Strategies in Seedling Nurseries
- Development and Application of Molecular Markers Associated with Anti-fusarium Wilt on Luffa and Drought-tolerance and Other Important Traits on Maize
- Study on Functional Microbiomes for Disease Resistance in Tomatoes
- Development of Key Technologies for Tissue Culture-assisted Seed Selection
- Development of Molecular Marker for Melon Resilience Breeding
- Research on germplasm maintenance and industrial application of vegetative propagation crops
- Germplasm Development and Application of Cattleya Alliance and Agapanthus.
- The Promotion of Aboriginal Beans Crops Seed Saving System
- Next-generation Ark of Agriculture and Forestry Germplasm - Germplasm Regeneration of Important Cross-Pollinated Crops.
- Establishment of Organic Seed Production and Supplying System for Agriculture and Hortculture Crops- Buckwheat, Tomato and Okra
- Techniques Development for Adaptable Varieties of Bulb Flowers Breeding
- Establishment of a Heat-Tolerance Selection Index and Breeding of Tomatoes
- High Quality and Stress Tolerance Breeding in Tomato.
- Breeding of New Papaya Varieties for Net House Production
- Research on the Development and Application of Diverse Food and Agriculture Teaching Materials of Potato
- Research on the Behavioral Intentions of Agricultural Social Responsibility
- Application Evaluation of Taro Diversified Processing Raw Materials
- Promote the Industrialization of Precise Strawberry Seedling Hardening Modules
- Enhancement of the Testing and Monitoring System for Genetically Modified Crops in the Central Region of Taiwan and the Import Channels
- Enhancing the Connection and Participation with Seed Testing Technology Related International Organization
 Home > Achievement > 2025 Research Project List > Establishment of Techniques for Phytoplasma Detection and Classification and Investigation on Phytoplasma Pathogen in Seeds of Important Crops Home > Achievement > 2025 Research Project List > Establishment of Techniques for Phytoplasma Detection and Classification and Investigation on Phytoplasma Pathogen in Seeds of Important Crops |
Establishment of Techniques for Phytoplasma Detection and Classification and Investigation on Phytoplasma Pathogen in Seeds of Important Crops
Phytoplasmas (Candidatus Phytoplasma) are among the most important plant pathogens worldwide and are known to infect more than a thousand plant species, including many fruit trees and economically valuable crops. They can spread through vegetative propagation methods such as cuttings and grafting, as well as through parasitic dodder and various insect vectors including leafhoppers, planthoppers, and psyllids. Infected plants often exhibit symptoms such as phyllody, stunting, yellowing, and witches’ broom, leading to substantial agricultural losses. Since phytoplasmas cannot be cultured in vitro, their detection and identification remain highly challenging. With increasing demands for plant quarantine and disease prevention, establishing internationally aligned detection and diagnostic methods for phytoplasmas is crucial for crop disease diagnosis and surveys of seed-associated pathogens in Taiwan. In the first year of this project, representative materials of the two major phytoplasma groups found in Taiwan were collected and established. These include the Periwinkle leaf yellowing (PLY) phytoplasma (16Sr I-B, aster yellows group) representing Group I (16Sr I) and the Peanut witches’ broom (PnWB) phytoplasma (16Sr II-A) representing Group II (16Sr II), collected from peanut fields in Tai-si and Tuku, Yunlin County. This material library provides an essential foundation for developing detection methods. Two nucleic acid extraction systems, TANBead and taco, were compared, and DNA purity was evaluated using A260/280 and A260/230 ratios. Results indicated that TANBead yielded higher DNA purity (1.79-1.91) and could be reliably applied to various suspected hosts, including Ixeris chinensis , Emilia sonchifolia , Arachis duranensis, and Cynodon dactylon and semi-nested PCR systems were established. The primer pair P1A/P7A was selected for the first PCR (approximately 1.8 kb), and P1A/16S-Sr for the semi-nested PCR (approximately 1.5 kb). Both PLY and PnWB phytoplasmas successfully produced the expected. Amplicons were sequenced, and the resulting sequences were compared using the BLAST tool in the NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/) database. Sequence identities were all above 99.2%, confirming that the samples correspond to PLY and PnWB phytoplasmas, respectively.
 ▲Figure 1. General Characteristics of Phytoplasmas (Candidatus Phytoplasma). |
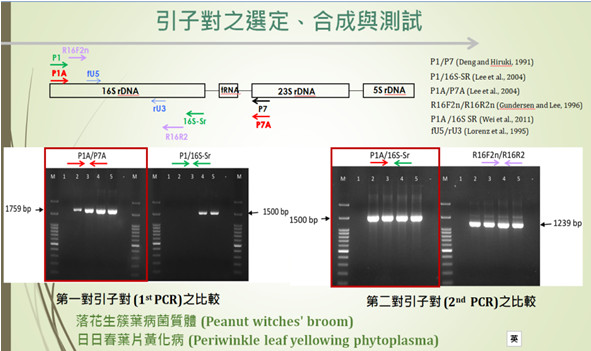 ▲Figure 2. Establishment of Detection Techniques for Phytoplasmas. |
