- Technology development of poultry feed for added value of by-products from tomato and pumpkin seed collection
- Establishment of upland crop waste mobile processing system
- Research on Green Manure Crops to Provide Dry Season nectar Source
- Establishing production adaptation and management techniques for drought-tolerant potato
- Research on Improving Agricultural Workers' Adaptation Strategies in Response to Climate Change: A Case Study of Agricultural Workers in Seedling production industries.
- Application of Herbal Medicine Plants in Animal Feed Additives and Establishment of GAP Production System.
- Constructing intelligent identification services for seed and seedling industry
- Construct expert system for forecasting the production of major important vegetables seedlings
- The establishment of smart production and application model of plant tissue culture
- The establishment of high through-put sequencing workflow to detect pathogens of tomato and Capsicum seeds
- Establishment of precise and efficient breeding technology for industry-oriented traits of cauliflower
- Molecular Marker-Assisted Tomato Disease Resistance Breeding Selection Model Construction and Technology Development
- Participatory selection of healthy ginger seedlings and establishment of production system
- Participatory selection of taro regional clonal lines
- Development of seed production technology for new grain crop varieties - Maize
- Establishment of highly effective isolated facility for healthy seedling
- Research of optimization for detecting operation procedure on seed-borne pathogens
- Study and improvement of strengthening seed testing technology
- Analysis and application of root‐associated microbes from soil-borne disease resistant and sensitive lines of Solanaceae crops
- Developing of crops novel assisted breeding technology on pepper and melon
- The study of healthy seedling production by in vitro micropropagation
- Germplasm development and application of Holcoglossum genus and Cattleya genus.
- Research on Genetic Resources and Plant New Variety DUS Testing Techniques Management and Application of the Economical and Special Crops in Taiwan
- Establishment of organic seed production and supplying system for grain crops
- The seedling leaf electrolyte leakage index was used to select papaya heat-resistant Resistance and virus Resistance varieties.
- Tomato hybrid seed miniature production
- Tomato(Solanaceae)resistance to bacterial wilt of selection in Field.
- Techniques development for bulb flowers breeding
- Breeding for disease resistant tomato and superior quality eggplant.
- Establish Cucurbitaceae vegetable breeding technologies of heat tolerance
- Establishing the Technology of New Papaya Viety Breeding for the International Market.
- Research on the Planning of Crop Seed Conservation Course and Training Performance Analysis for The Seedling Training Courses
- Establish a digital platform for seed illustration
- Establishment of automatic seedling evaluation system and integration of seed testing platform
- Study of inspection and monitoring system for precisely breeding crops
- Enhancing the seed testing technical cooperation with ISTA and new southbound countries
 Home > Achievement > 2021 Research Project List > The establishment of high through-put sequencing workflow to detect pathogens of tomato and Capsicum seeds Home > Achievement > 2021 Research Project List > The establishment of high through-put sequencing workflow to detect pathogens of tomato and Capsicum seeds |
The establishment of high through-put sequencing workflow to detect pathogens of tomato and Capsicum seeds
Using small molecule RNA(sRNAs) as a detect target to clarify the consortia and identities of pathogens in import and export of tomato seeds. Specifically, a CLC NGS analysis system based on LINUX will be built up. Small RNAs from 5 tomato samples will be extracted and used for sequencing. The raw data of these sequences will be processed using a serial of bioinformatics tools and programs. Then an analysis workflow for deconstructing sequencing data into plant pathogen identities and a sRNA database of tomato host plant will be set up. Along with hardware updating with high-performance computing processors, this workflow will help us to establish sRNA detection procedures for tomato pathogens. Moreover, batched of import and export tomato seeds will be collected and tested. The purpose of this project is to collect and analyze small molecule RNA sequence data to establish a method for plant pathogen detection. Assisting domestic seed and seedling operators to verify the pathogens of important materials and to establish a source database of healthy species. This technology can be applied to applications for new crop varieties. Samplers can go to the exporting country to investigate the epidemic materials. Border guards can also conduct disease phase analysis for centralized input samples.
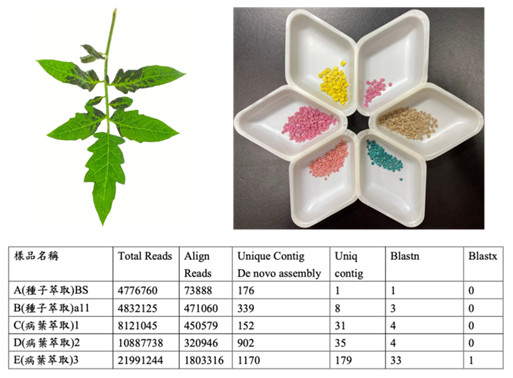 ▲Fig. 1. The small RNA sequencing results of seed and leaf samples of tomato and Capsicum. |
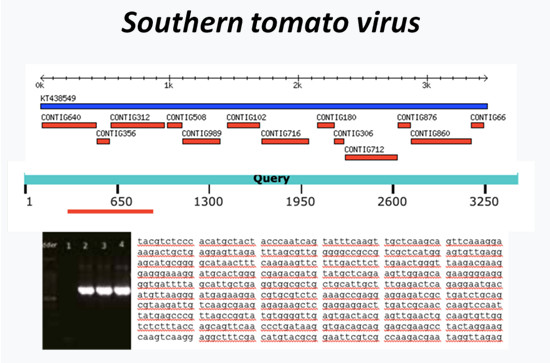 ▲Fig. 1. Using RT-PCR and amplicon sequencing to confirm digital analysis results of the small RNA sequencing data. |
