- Research on Seedling Propagation Technology of Eastern Aboriginal Tribes
- Counseling on Digital Transformation of Small,Medium and Micro-sized Crops and Seedlings with Potential for Export
- Recycling of surplus materials from tomato and pumpkin productio
- Research on the improvement of a renewable energy supply chain model for the residual media of mushroom
- Adaptation and mitigation the impact of important weeds on staple crops (cucumber) under climate change
- Constructing intelligent identification services for seed and seedling industry
- Application and Promotion of Production and Sales Management Wisdom Network for Vegetable Nursery
- Function Demonstration and Optimization of the Expert System for Vegetable Seedling Production Forecast
- The establishment of smart production and application model of plant tissue culture
- Research of optimization for detecting operation procedure on seed-borne pathogens and development of seed disinfection technique
- Research and consulting on the response of seed industry under high temperature and drought stress.
- Establishment of the heat-resistant selection index and breeding of tomato
- High quality tomato and stress tolerance breeding technology in Facility.
- Evaluation and Application of Drought-tolerant Potato Germplasm Resources
- Establishment of the biological indicator of disease resistance for tomato in drought condition
- Establishment of marker assisted selection applied to the selection of drought-tolerant maize and heattolerant tomato
- Establishment of precise and efficient breeding technology for industry-oriented traits of cauliflower
- Molecular Marker-Assisted Tomato Disease Resistance Breeding Selection Model Construction and Technology Development
- Participatory Selection of Healthy Ginger Seedlings and Establishment of Production System
- Participatory selection of taro regional clonal lines
- Analysis and application of root‐associated microbes from soil-borne disease resistant and sensitive lines of Solanaceae crops
- Improvement on the Micropropagation Technique - In vitro propagation of avocado (Persea americana Mill.) through apical buds and nodal segments
- Developing of crops novel assisted breeding technology on melon
- Research on germplasm maintenance and industrial application of vegetative propagation crops
- Germplasm development and application of Aeridinae subtribe orchids and Cattleya genus.
- The Establishment of Organic Seedling and Seed Saving System
- Research on Genetic Resources and Plant New Variety DUS Testing Techniques Management and Application of the Economical and Special Crops in Taiwan
- Establishment of organic seed production and supplying system for grain crops
- The seedling leaf electrolyte leakage index was used to select papaya heat-resistant Resistance and virus Resistance varieties.
- Selecting for resistance bacterial wilt and F1 Seed Collection of tomato.
- Techniques development for bulb flowers breeding
- Establish Cucurbitaceae vegetable breeding technologies of heat tolerance
- Establishing the Technology ofNew Papaya Viety Breeding for the International Market.
- Research on Agricultural Social Responsibility for the trainees of Farmers' Academy
- Establishment of Phalaenopsis Varieties Identification and Application Integration Platform
- Study of inspection and monitoring system for precisely breeding crops
- Harmonization and Cooperation on DUS testing between Taiwan and Japan
- Taiwan-Israel Plant Variety Protection Cooperation and Study on DUS Testing
 Home > Achievement > 2022 Research Project List > Analysis and application of root‐associated microbes from soil-borne disease resistant and sensitive lines of Solanaceae crops Home > Achievement > 2022 Research Project List > Analysis and application of root‐associated microbes from soil-borne disease resistant and sensitive lines of Solanaceae crops |
Analysis and application of root‐associated microbes from soil-borne disease resistant and sensitive lines of Solanaceae crops
Tomato soil-borne diseases may cause around 10-20% yield loss annually. Due to intensive cultivation and climate change, infectious cases are increasing. Recent studies have shown that adjusting root microbes may benefit the crops to become more resilient to environmental change. Therefore, this project aims to conduct a metagenomics analysis of the cultivation soil from tomato root-knot nematode-resistant and susceptible genotype lines. By analyzing the differences between the root microbiome, we wish to screen possible molecular targets and isolate the core microbes which could benefit the tomato growth. At present, the sequence analysis of 16S rDNA amplicons from soil samples has been conducted. The OUT heatmap and Venn diagram have been performed to reveal the abundance of OTU as well as the similarity and difference across samples. At the same time, box plots, unifrac distance matrix, PCA analysis, Anosim analysis, Metastats, and LefSe analysis were performed to compare the diversity and differences between groups. Moreover, the isolation of rhizosphere microbes from root-knot nematode resistant genotype tomato plants was also conducted.
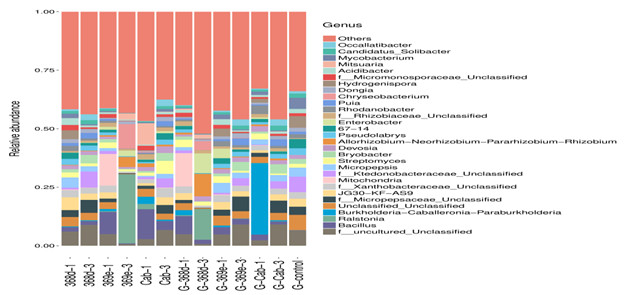 ▲Fig.1. Comparison of the soil community structures in the mesocosm experiment based on pyrosequencing of 16S rDNA amplicons. |
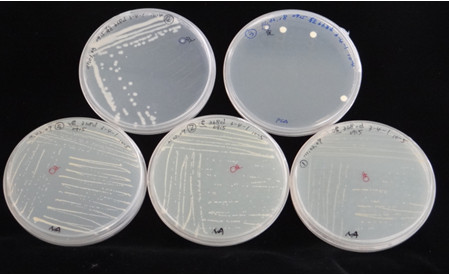 ▲Fig.2 Isolation of rhizosphere microorganisms from root-knot nematode resistant genotype tomato plants. ▲Fig.2 Isolation of rhizosphere microorganisms from root-knot nematode resistant genotype tomato plants. |
